Human Migration and Ancient DNA
Pedersen MW, Antunes C, De Cahsan B, Moreno-Mayar JV, Sikora M, Vinner L, Mann D, Klimov PB, Black S, Michieli CT, Braig HR, & Perotti MA. 2022
Ancient human genomes and environmental DNA from the cement attaching 2,000 year-old head lice nits.
Molecular Biology and Evolution 39, msab351.
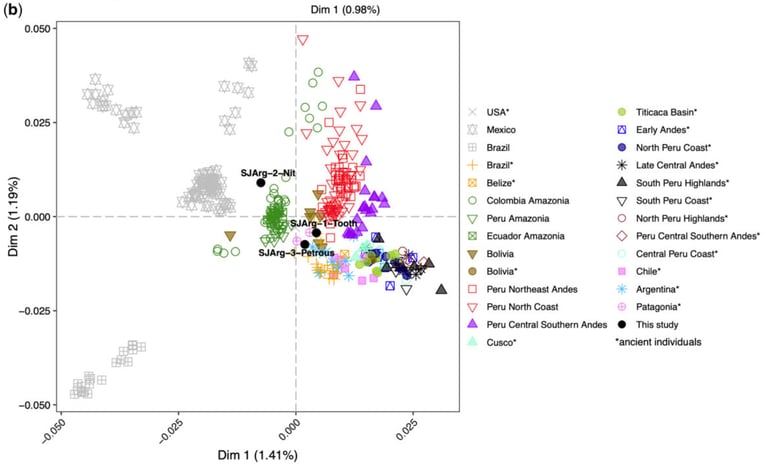
Over the past few decades, there has been a growing demand for genome analysis of ancient human remains. Destructive sampling is increasingly difficult to obtain for ethical reasons, and standard methods of breaking the skull to access the petrous bone or sampling remaining teeth are often forbidden for curatorial reasons. However, most ancient humans carried head lice and their eggs abound in historical hair specimens. Here we show that host DNA is protected by the cement that glues head lice nits to the hair of ancient Argentinian mummies, 1,500–2,000 years old. The genetic affinities deciphered from genome-wide analyses of this DNA inform that this population migrated from north-west Amazonia to the Andes of central-west Argentina; a result confirmed using the mitochondria of the host lice. The cement preserves ancient environmental DNA of the skin, including the earliest recorded case of Merkel cell polyomavirus. We found that the percentage of human DNA obtained from nit cement equals human DNA obtained from the tooth, yield 2-fold compared with a petrous bone, and 4-fold to a bloodmeal of adult lice a millennium younger. In metric studies of sheaths, the length of the cement negatively correlates with the age of the specimens, whereas hair linear distance between nit and scalp informs about the environmental conditions at the time before death. Ectoparasitic lice sheaths can offer an alternative, nondestructive source of high-quality ancient DNA from a variety of host taxa where bones and teeth are not available and reveal complementary details of their history.
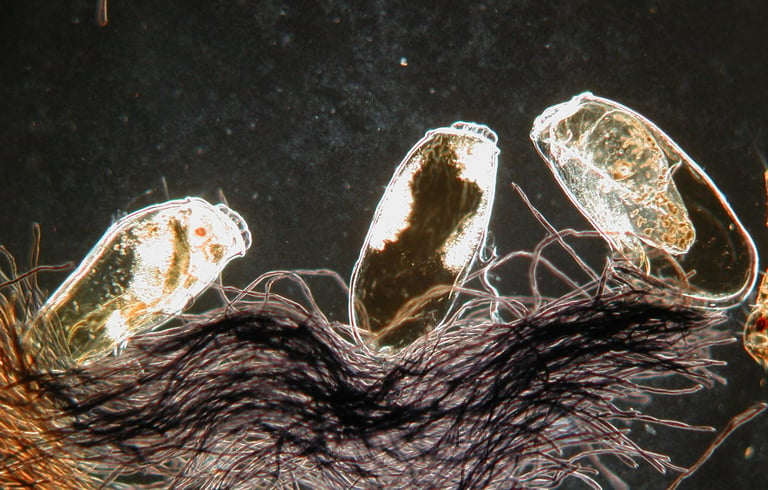
Perotti MA, Reed D, Mumcuoglu KY, Pedersen MW, Klimov PB, & Braig HR. 2021
Human lice as mirrors and archives of host evolutionary history – Lice offer host DNA via non-destructive sampling.
Cambridge, United Kingdom: Human Evolution – From Fossils to Ancient and Modern Genomes, 02–04 November 2021, Wellcome Genome Campus Conference.
Among the ectoparasites of humans – fleas, parasitic flies, ticks, and mites – the lice stand out. Along a few mite species, they are the only ectoparasites that cannot survive off the host for more than one or two days. They are transmitted from person to person through direct contact. If the host dies, they die with their host. Unlike most insects, lice cannot fly. They are obligatory bloodsucking insects that have distinct niches upon humans, either on the scalp, head lice, Pediculus humanus capitis, inside clothing, body lice, P. humanus humanus, or on pubic and eyebrow hairs, Pthirus pubis. The three nymphal stages and adult male and female lice require bloodmeals. Female head lice live for up to five weeks and produce up to three eggs per day, while body lice lay twice this number.
Archaic and early modern humans were supposedly very lousy. Because of the intense infestations in the past, ancient human remains or sediments associated with human habitation or exploitation almost always carry louse remains. On the hosts, they are found mainly attached to hairs or garments as nits, which are the eggs and embryos encased in a protective structure glued to hairs or fibres. Since humans started using combs like today’s louse combs, they also collect nits on these combs. In sediments, remains of hair with nits attached and loose adult lice are also very frequent. Despite being so prevalent, they are often overlooked from archaeological material such as skeletons, hair, combs, and mummies and their textiles.
Lice carry host DNA that can be of the same quality as DNA extracted from a petrous bone or a tooth, offering unique details of host genetic affinities while preventing destruction of valuable archaeological specimens. Lice from forensic and archaeological samples are reliable witnesses of the environment their hosts were living in, of the circumstances around the life and death, and the health status of the hosts. They even inform of pathogens carried by them as vectors or by their infected hosts.
Contact between contemporaneous human lineages results in the exchange of lice lineages, making these lice and their DNA sequence data a very informative tool for inquiring the hosts' evolutionary past and history.
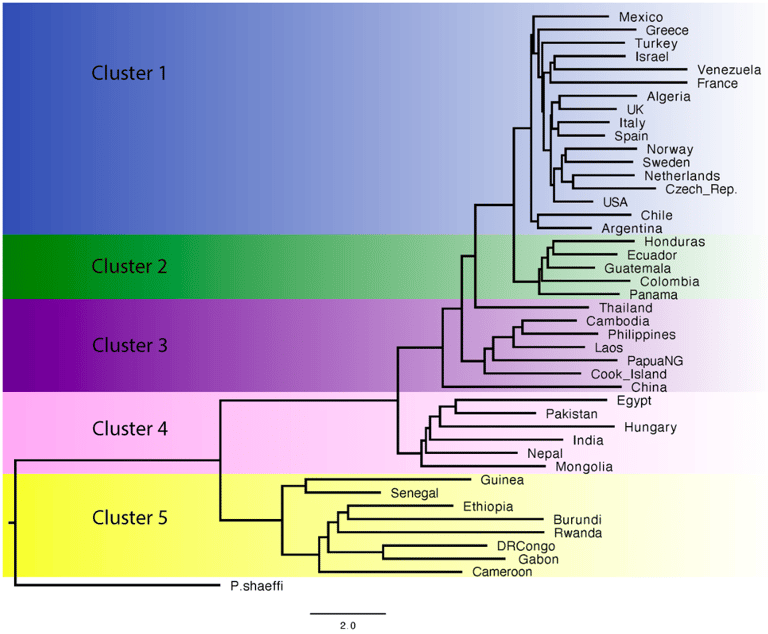
House N, Miro-Herrans AT, Allen JM, Ashfaq M, Boyd BM, Braig HR, Mediannikov O, Mehlhorn H, Mumcuoglu KY, Perotti MA, Ramíres Rozzi F, Raoult D, Rozsa L, Stefka J, Toloza A C, Wirdemark NV, & Reed DL. 2024
Genome-wide analysis of the human head louse (Pediculus humanus capitis) reveals geographically structured genetic populations.
Authorea preprint, under review.
The human head louse (Pediculus humanus capitis) is an obligate ectoparasite of humans and has the potential to uncover aspects of human history that cannot be directly inferred from genetic data derived from humans. Previous studies have shown that global louse populations exhibit restricted patterns of genetic variation. However, these studies were restricted both genetically and lacked a global sampling. With the aim of capturing the genetic diversity of head louse populations from around the world, we generated whole genome sequences of human head lice from 43 countries, spanning five continents and Oceania, to determine if louse nuclear diversity mirrors its mitochondrial haplotypes or if population genetic structure, genetic diversity, and population connectivity are associated with geographical regions or host behavior. Here we show that there are five nuclear genetic clusters that are associated with large geographical regions, either at continental or intercontinental levels. High genetic variation was found between African and non-African individuals and the highest genetic diversity was found in samples from sub-Saharan Africa, similar to that of humans. Unlike the mitochondrial clades examined in previous studies, nuclear genetic clusters of lice examined here are highly structured based on geography (continentally and major regions within continents). Results from our genome analyses revealed that host-mediated global dispersal as the likely primary process in shaping diversity and maintaining genetic population boundaries within the nuclear genome of the human head louse.